AKUNA FAVOUR 18MHS06012 MED LAB SCI REPLICATION OCCUR IN
AKUNA FAVOUR 18MHS06012 MED LAB SCI REPLICATION OCCUR IN
AKUNA FAVOUR
18/MHS06/012
MED LAB SCI
replication occur in 3 steps
Initiation,
Elongation,
Termination
1.Initiation
DNA synthesis is initiated at particular points within the DNA strand known as ‘origins’, which are specific coding regions. These origins are targeted by initiator proteins, which go on to recruit more proteins that help aid the replication process, forming a replication complex around the DNA origin. There are multiple origin sites, and when replication of DNA begins, these sites are referred to as Replication Forks.
Within the replication complex is the enzyme DNA Helicase, which unwinds the double helix and exposes each of the two strands, so that they can be used as a template for replication. It does this by hydrolysing the ATP used to form the bonds between the nucleobases, therefore breaking the bond between the two strands.
DNA can only be extended via the addition of a free nucleotide triphosphate to the 3’- end of a chain. As the double helix runs antiparallel, but DNA replication only occurs in one direction, it means growth of the two new strands is very different (and will be covered in Elongation).
DNA Primase is another enzyme that is important in DNA replication. It synthesises a small RNA primer, which acts as a ‘kick-starter’ for DNA Polymerase.DNA Polymerase is the enzyme that is ultimately responsible for the creation and expansion of the new strands of DNA.
2.Elongation
Once the DNA Polymerase has attached to the original, unzipped two strands of DNA (i.e. the template strands), it is able to start synthesising the new DNA to match the templates. This enzyme is only able to extend the primer by adding free nucleotides to the 3’-end of the strand, causing difficulty as one of the template strands has a 5’-end from which it needs to extend from.
One of the templates is read in a 3’ to 5’ direction, which means that the new strand will be formed in a 5’ to 3’ direction (as the two strands are antiparallel to each other). This newly formed strand is referred to as the Leading Strand. Along this strand, DNA Primase only needs to synthesise an RNA primeronce, at the beginning, to help initiate DNA Polymerase to continue extending the new DNA strand. This is because DNA Polymerase is able to extend the new DNA strand normally, by adding new nucleotides to the 3’ end of the new strand (how DNA Polymerase usually works).
However, the other template strand is antiparallel, and is therefore read in a 5’ to 3’ direction, meaning the new DNA strand being formed will run in a 3’ to 5’ direction. This is an issue as DNA Polymerase doesn’t extend in this direction. To counteract this, DNA Primase synthesises a new RNA primer approximately every 200 nucleotides, to prime DNA synthesis to continue extending from the 5’ end of the new strand. To allow for the continued creation of RNA primers, the new synthesis is delayed and is such called the Lagging Strand.
The leading strand is one complete strand, while the lagging strand is not. It is instead made out of multiple ‘mini-strands’, known of Okazaki fragments. These fragments occur due to the fact that new primers are having to be synthesised, therefore causing multiple strands to be created, as opposed to the one initial primer that is used with the leading strand.
3.Termination
The process of expanding the new DNA strands continues until there is either no more DNA template left to replicate (i.e. at the end of the chromosome), or two replication forks meet and subsequently terminate. The meeting of two replication forks is not regulated and happens randomly along the course of the chromosome.
Once DNA synthesis has finished, it is important that the newly synthesised strands are bound and stabilized. With regards to the lagging strand, two enzymes are needed to achieve this; RNAase Hremoves the RNA primer that is at the beginning of each Okazaki fragment, and DNA Ligase joins two fragments together creating one complete strand.
Now with two new strands being finally finished, the DNA has been successfully replicated, and will just need other intrinsic cell systems to ‘proof-read’ the new DNA to check for any errors in replication, and for the new single strands to be stabilized.
[5/6, 9:41 PM] Seun: Important Enzymes in DNA Replication
Enzyme Function
Topoisomerase Relaxes the super-coiled DNA
DNA helicase Unwinds the double helix at the replication fork
Primase Provides the starting point for DNA polymerase to begin synthesis of the new strand
DNA polymerase Synthesizes the new DNA strand; also proofreads and corrects some errors
DNA ligase Re-joins the two DNA strands into a double helix and joins Okazaki fragments of the lagging st
Tags: akuna, replication, favour, occur, 18mhs06012
- (IMIĘ I NAZWISKO STUDENTA) (ROK KIERUNEK SYSTEM
- EDWARD HALL KULTURÁLIS KONCEPCIÓI KONTEXTUS KÖZÖS JELENTÉSTARTALMI HÁTTÉR
- FORMULARZ ZGŁOSZENIA ODBIORU ZSEE NAZWA FIRMY ULICA KOD MIEJSCOWOŚĆ
- ZAŁĄCZNIK DO UCHWAŁY RADY MIASTA NR XLIV41305 Z DNIA
- ESPAÑOL 1SS NOMBRE CUENTO EN DIBUJOS FECHA PERÍODO REFLEJA
- FORMULARIO FI006 FORMULARIO DE QUEJASRECLAMOSUGERENCIA 1 DATOS DE
- S UNDS GF SAMARBEJDSUDVALGET (SU) ARBEJDE OG FORRETNINGSORDEN 20212022
- MEMO TO DEBT MANAGEMENT OFFICE TOTAL DUE EXCEEDS
- (IMIĘIMIONA I NAZWISKO) (WYDZIAŁ) (KIERUNEK ROK
- COLEGIO CRISTÓBAL COLÓN MATEMÁTICA Iº MEDIO PROF ROMINA MONSALVE
- FACULTAD DE GEOGRAFÍA E HISTORIA INFRAESTRUCTURAS EQUIPAMIENTOS Y SERVICIOS
- ENPRESAIDEIA LEHIAKETA CONCURSO DE IDEAS DE NEGOCIO PARTEHARTZEKO ESKABIDE
- GRADE 6 MODULE 2A UNIT 1 LESSON 10 GETTING
- ¡¡TE INVITAMOS A NUESTRO DIA DEL NIÑO!! EL PRÓXIMO
- Vortrag Teresa Schweiger 23 Juni 2009 Sprache und Identität
- OSTRÓW WIELKOPOLSKI DNIA ………………………………………… ………………………………………… ………………………………………… DANE WNIOSKODAWCY
- TRASFERIMENTO PER ATTO TRA VIVI TAXINCC AL COMUNE DI
- EL NUEVO CERTAMEN RECUPERA ‘MARGARITA Y EL LOBO’ CINTA
- BOULACHIN PIERREANDRÉ GI ANALYSE SYSTÉMIQUE LE CAS BÉNÉTEAU
- CONCEPTO Y CLASIFICACIÓN CON EL NOMBRE DE LÍPIDOS (DEL
- NÁZOV PREDMETU RUSKÝ JAZYK (1 CUDZÍ JAZYK) ČASOVÝ ROZSAH
- 8 MEĐUNARODNA FILMSKA MANIFESTACIJA „UHVATI SA MNOM OVAJ DAN!“
- TÍTULO “ASTRONOMÍA Y ASTROBIOLOGÍA CON INTERNET RECURSOS INFORMÁTICOS Y
- NAZWA I ADRES LABORATORIUM1) ZLB3 ZGŁOSZENIE DODATNIEGO WYNIKU BADANIA
- COLEGIO SANTA ROSA DEPARTAMENTO DE MATEMÁTICAS BAYAMÓN PUERTO RICO
- EJEMPLO DE ARCHIVO ADJUNTO A UNA NOTICIA
- 精神科常見用藥 20116友靖老師製 商品名(劑量) 學名 藥理機轉 副作用 注意事項(外觀敘述) ATIVAN(ANXICAM) LORAZEPAM
- PROSJEKTBESKRIVELSE FOR VIDEREFØRING AV BRUKERFORUM I STANGE OG HAMAR
- FORMULARZ ZGŁOSZENIOWY (OSOBA UCZĄCA SIĘ) DO PROJEKTU PN „HISZPAŃSKIE
- COLEGIO EL VALLE EJERCICIOS DE VERANO 3ºESO TRABAJO DE
2 PRIKAZ STANJA PROSTORA KAZALO 1 UVOD 2 2
VISTO EL CONTRATO DE CONCESIÓN PARA LA PRESTACIÓN DEL
LICENSE AGREEMENT FOR AN INDEXARB INSTITUTIONAL SERVICES SUBSCRIPTION
DEPARTAMENTO DE BAJA VISION REHABILITACION CUAL ES EL OBJETIVO?
“NGÀY HỘI GIA ĐÌNH VIỆT NAM NĂM 2014”
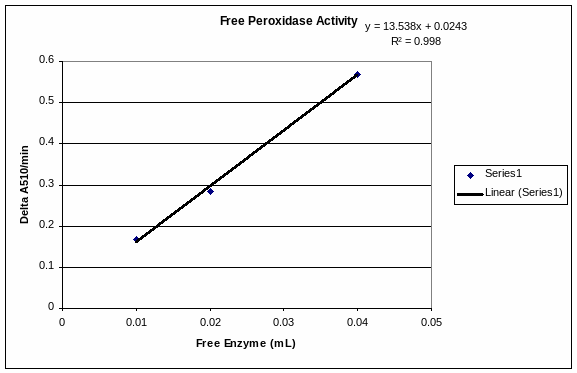 ACTIVITY AND THERMAL STABILITY OF GELIMMOBILIZED PEROXIDASE AUTHOR JASON
ACTIVITY AND THERMAL STABILITY OF GELIMMOBILIZED PEROXIDASE AUTHOR JASON SØKNAD OM MOTORFERDSEL I UTMARK LEGEERKLÆRING AJOUR PR 10
SØKNAD OM MOTORFERDSEL I UTMARK LEGEERKLÆRING AJOUR PR 10CONFORMITY DEFINITIONS SOCIAL NORMS IMPLICIT OR EXPLICIT RULES FOR
ADDENDUM (SOUTH DAKOTA) HUD PROJECT NUMBER PROJECT NAME A
 ANEXO BI SOLICITUD DE APROBACIÓN DEL PROYECTO Y
ANEXO BI SOLICITUD DE APROBACIÓN DEL PROYECTO YTC YARGITAY HUKUK GENEL KURULU E 20105662 K 2010651
COMMITTEE ON TRADE AND INVESTMENT SUMMARY RECORD OF THE
MODELO DE DECLARACION JURADA DEL PRESIDENTE DE LA ASOCIACION
 ERCÜMENT KAVAK DOĞUM TARIHI 11 ARALIK 1970 MEDENI DURUMU
ERCÜMENT KAVAK DOĞUM TARIHI 11 ARALIK 1970 MEDENI DURUMU [TYPE TEXT] RIVERSIDE ESTATES TENANCY LEASE AGREEMENT NOVA SCOTIA
[TYPE TEXT] RIVERSIDE ESTATES TENANCY LEASE AGREEMENT NOVA SCOTIAXXX CONGRESO ALAS 2015 COSTA RICA PONENCIA MUJER Y
 D UNAALMÁS KÖZSÉG POLGÁRMESTERÉTŐL 2545 DUNAALMÁS ALMÁSI U 32
D UNAALMÁS KÖZSÉG POLGÁRMESTERÉTŐL 2545 DUNAALMÁS ALMÁSI U 32TATO ŽÁDOST BYLA JIŽ PROJEDNÁNA A SCHVÁLENA NA VALNÉ
MARTES 15 DE JUNIO DE 2010 DIARIO OFICIAL (CUARTA
JURY OF 1ST ROUND EU PARTNERS LNU DR OLA