SUPPLEMENTAL INFORMATION A MICROFLUIDIC CHIP FOR DETECTING CHOLANGIOCARCINOMA CELLS
1 SUPPLEMENTAL TABLE THE EMPYREAN STUDY COMMITTEES PRINCIPAL INVESTIGATOR11 SUPPLEMENTAL FIGURES SF1 SAMPLES CLUSTER BY STRAIN PLOT
2170463 §217046—ELIGIBILITY FOR SUPPLEMENTAL EDUCATIONAL ASSISTANCE 2170463 §217046 ELIGIBILITY
3 SUPPLEMENTAL TABLE 1 PARTIAL CORRELATIONS† BETWEEN DIETARY FACTORS‡
7 SUPPLEMENTAL INFORMATION VAKIFAHMETOGLUNORBERG ET AL SUPPLEMENTAL INFORMATION SUPPLEMENTAL
7 TABLE 1 SUPPLEMENTAL INFORMATION ON THE ANALYZED STUDIES
Supplemental information
A MICROFLUIDIC CHIP FOR DETECTING CHOLANGIOCARCINOMA CELLS IN HUMAN BILE*
Lien-Yu Hung1†, Nai-Jung Chiang2,3‡, Wei-Chun Tsai1, Chien-Yu Fu1, Yu-Chun Wang4, Yan-Shen Shan4+, and Gwo-Bin Lee1,5,6*
1Department of Power Mechanical Engineering, National Tsing Hua University, Hsinchu, Taiwan
2National Institute of Cancer Research, National Health Research Institutes, Tainan, Taiwan
3Division of Hematology and Oncology, Department of Internal Medicine, National Cheng Kung University Hospital, Tainan, Taiwan
4Institute of Clinical Medicine, National Cheng Kung University Hospital, Tainan, Taiwan
5Institute of Biomedical Engineering, National Tsing Hua University, Hsinchu, Taiwan
6Institute of NanoEngineering and Microsystems, National Tsing Hua University, Hsinchu, Taiwan
†First author: Dr. Lien-Yu Hung; email: [email protected].
‡Co-first author: Dr. Nai-Jung Chiang; email: [email protected].
*Corresponding author: Dr. Gwo-Bin Lee; email: [email protected]; Tel: +886-3-5711531 ext. 33765; Fax: +886-3-5742495
+Co-corresponding author: Dr. Yan-Shen Shan; email: [email protected]; Tel: +886-6-2353535 ext. 3105; Fax: +886-6-2758781
The experimental protocol of the on-chip Cell-SELEX
Supplemental Table 1: detailed information about the experimental protocol on the Cell-SELEX microfluidic platform
|
Step |
Procedure |
Sample volume (μL) |
On-chip operation condition |
|
1. |
Load anti-EpCAM beads and pre-treated bile into the chamber A |
10/600 |
|
|
|
Load 1×PBS into the chambers P |
600 |
|
|
|
Load 16% paraformaldehyde into the chamber D |
200 |
|
|
|
Load 0.1% Triton X 100 into the chamber E |
200 |
|
|
|
Load diluted first antibody chamber into the chamber F |
200 |
|
|
|
Load diluted secondary antibody into the chamber G |
200 |
|
|
|
Load diluted DAPI/Hoechst into the chamber H |
200 |
|
|
2. |
Incubate anti-EpCAM beads and pre-treated bile for 15 min |
|
-100 mmHg and 0.5 Hz for the micromixer |
|
3. |
Collected bead-cancer cell complexes by a magnet and remove the incubation supernatant through the waste chamber |
|
-500 mmHg for the suction pressure |
|
4. |
Transport 1×PBS into the micromixer |
600 |
-100 mmHg and 0.5 Hz for the micromixer |
|
5. |
Wash the collected bead-cancer cell complexes by the micromixer for 1 min |
|
-100 mmHg and 0.5 Hz for the micromixer |
|
6. |
Remove the incubation supernatant through the waste chamber |
|
-500 mmHg for the suction pressure |
|
7. |
Repeat the steps 4-6 for another two times |
|
|
|
8. |
Transport 1×PBS into the micromixer |
600 |
-100 mmHg and 0.5 Hz for the micromixer |
|
9. |
Incubate bead-cancer cell complexes with 4% paraformaldehyde for 3 min |
|
-100 mmHg and 0.5 Hz for the micromixer |
|
10. |
Collected bead-cancer cell complexes by a magnet and remove the incubation supernatant through the waste chamber |
|
-500 mmHg for the suction pressure |
|
11. |
Transport 1×PBS into the micromixer and wash the collected bead-cancer cell complexes by the micromixer for 1 min |
600 |
-100 mmHg and 0.5 Hz for the micromixer |
|
12. |
Repeat the steps 10-11 for another two times |
|
|
|
13. |
Collected bead-cancer cell complexes by a magnet and remove the incubation supernatant through the waste chamber |
|
-500 mmHg for the suction pressure |
|
14. |
Transport 1×PBS into the micromixer |
600 |
-100 mmHg and 0.5 Hz for the micromixer |
|
15. |
Incubate bead-cancer cell complexes with 0.1% Triton X 100 for 3 min |
|
-100 mmHg and 0.5 Hz for the micromixer |
|
16. |
Collected bead-cancer cell complexes by a magnet and remove the incubation supernatant through the waste chamber |
|
-500 mmHg for the suction pressure |
|
17. |
Transport 1×PBS into the micromixer and wash the collected bead-cancer cell complexes by the micromixer for 1 min |
600 |
-100 mmHg and 0.5 Hz for the micromixer |
|
18. |
Repeat the steps 16-17 for another two times |
|
|
|
19. |
Collected bead-cancer cell complexes by a magnet and remove the incubation supernatant through the waste chamber |
|
-500 mmHg for the suction pressure |
|
20. |
Transport 1×PBS into the micromixer |
600 |
-100 mmHg and 0.5 Hz for the micromixer |
|
21. |
Incubate diluted first antibody and pre-treated bile for 15 min |
|
-100 mmHg and 0.5 Hz for the micromixer |
|
22. |
Collected bead-cancer cell complexes by a magnet and remove the incubation supernatant through the waste chamber |
|
-500 mmHg for the suction pressure |
|
23. |
Transport 1×PBS into the micromixer and wash the collected bead-cancer cell complexes by the micromixer for 1 min |
600 |
-100 mmHg and 0.5 Hz for the micromixer |
|
24. |
Repeat the steps 22-23 for another two times |
|
|
|
25. |
Collected bead-cancer cell complexes by a magnet and remove the incubation supernatant through the waste chamber |
|
-500 mmHg for the suction pressure |
|
26. |
Transport 1×PBS into the micromixer |
600 |
-100 mmHg and 0.5 Hz for the micromixer |
|
27. |
Incubate diluted secondary antibody and diluted DAPI/Hoechst pre-treated bile for 3 min |
|
-100 mmHg and 0.5 Hz for the micromixer |
|
28. |
Collected bead-cancer cell complexes by a magnet and remove the incubation supernatant through the waste chamber |
|
-500 mmHg for the suction pressure |
|
29. |
Transport 1×PBS into the micromixer and wash the collected bead-cancer cell complexes by the micromixer for 1 min |
600 |
-100 mmHg and 0.5 Hz for the micromixer |
|
30. |
Repeat the steps 28-29 for another two times |
|
|
|
31. |
Collected bead-cancer cell complexes by a magnet and remove the incubation supernatant through the waste chamber |
|
-500 mmHg for the suction pressure |
|
32. |
Transport 1×PBS into the micromixer |
600 |
-100 mmHg and 0.5 Hz for the micromixer |
|
33 |
Collected all re-suspended bead-cancer cell complexes for further fluorescence microscopy analysis. |
|
|
Characterization of the micromixer/micropumps, and microvalves
The integrated microfluidic chip was consisted of three layers, including two PDMS layers and one glass substrate. The first PDMS layer was a thin layer with a thickness of 150 μm, which was a liquid channel layer. Another thick PDMS layer (around 1500 μm), which contained air channels, was used to control the micropumps.
The main function of the transportation unit of the microfluidic chip was to transport the binding buffer and the washing buffer to the target cell region. Because the transport process requires three steps, reducing the operating time for each fluid transport step could be effectively achieved by increasing the overall flow rate. The fluidic pumping rate was also found to increase with an increase in the applied gauge pressure (i.e. higher vacuum). The maximum pumping rate of the transportation unit was found to be 233.5 μL/sec when operated at a frequency of 0.5 Hz at a gauge pressure of -100 mmHg.
The on-chip CCA cancer cell capture protocol which required pre-processing of bile samples with spiked-in cells
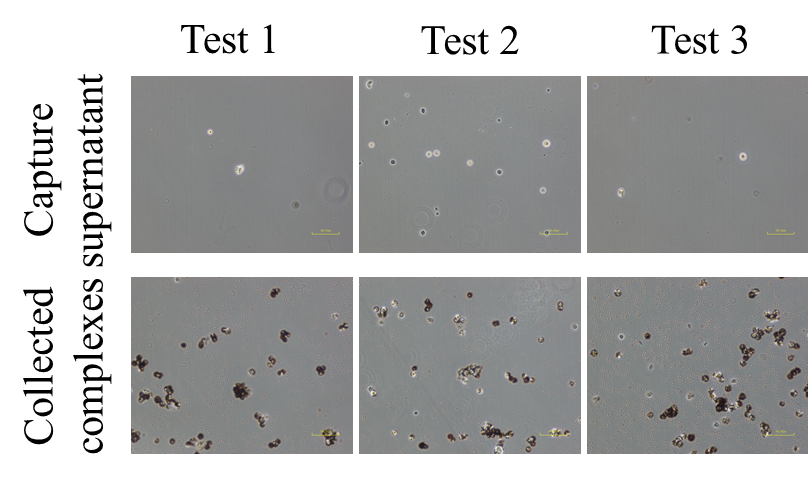
The on-chip CCA cancer cell capture protocol which required pre-processing of bile samples were performed with large (2×105) and small amounts (10 cells and one cell) of spiked-in cells, which presented 82.5±4.3, 70.0±8.1, and 66.6±4.7%, respectively, as shown in Supplemental Information Figure 2, Figure 3, Supplemental Information Table 2, and Table 3. It is important to note that these cell-spiked bile fluids were purified from diseased-bile samples by centrifugation since there was no non-diseased bile sample available.
Supplemental Information Figure 1. The on-chip CCA cancer cell capture protocol which required pre-processing of bile samples was performed with a relatively large amount of spiked-in cells (2×105)
Supplemental Information Table 2. Counted cells with a relatively large amount of spiked-in cells (2×105)
|
Spike-in cells |
Test 1 |
Test 2 |
Test 3 |
Capture rate (%) |
|
2×105 |
1.75×105 |
1.70×105 |
1.50×105 |
82.5±4.3 |
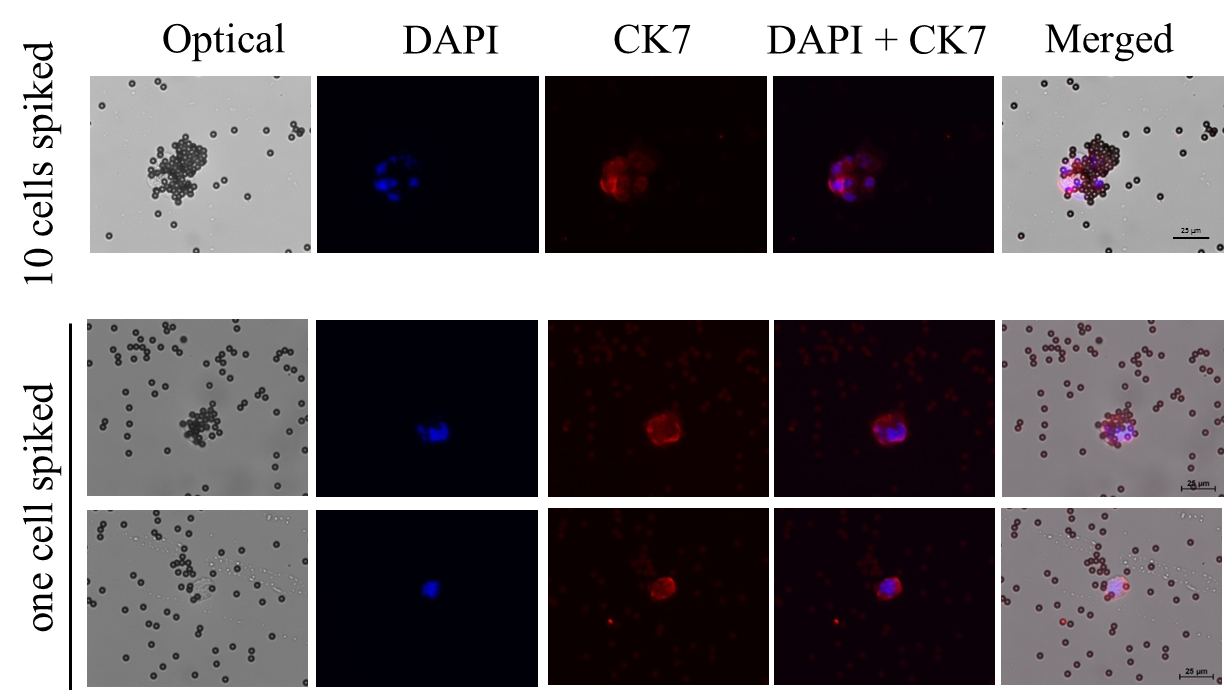
Supplemental Information Figure 2. The on-chip CCA cancer cell capture protocol which required pre-processing of bile samples was performed with a small amount of spiked-in cells (10 cells and one cell).
Supplemental Information Table 3. Counted cells with a small amount of spiked-in cells (10 cells and one cell)
|
Spike-in cells |
Test 1 |
Test 2 |
Test 3 |
Capture rate (%) |
|
10 |
6 |
8 |
7 |
70.0±8.1 |
|
1 |
1 |
1 |
0 |
66.6±4.7 |
The fluorescent images presented under bench-top IF staining process

Supplemental Information Figure 3. fluorescent images presented under bench-top IF staining process.
Achievement Goals 7 Supplemental Materials in Their own Words
ANNEX II SUPPLEMENTAL STATUTORY DECLARATION IN THE MATTER
ANNOUNCEMENT REQUEST FOR PROPOSAL GROUP LIFEDISABILITYSUPPLEMENTAL LIFE DECEMBER 2017
Tags: cells in, spike-in cells, cholangiocarcinoma, cells, information, microfluidic, supplemental, detecting
- PLAINS NATIVE AMERICANS THE PLAINS PEOPLE BLACKFOOT BLACKFOOT INDIANS
- VIDEO TRAINING PRESSURE ULCER PREVENTION ANSWERS (15)
- KERTAS KERJA MENGANJURKAN PERTANDINGAN NASYID ANTARA SEKOLAHSEKOLAH RENDAH DAERAH
- NOVEMBER 2014 BEANS PANTRY STAPLES NUTRITION STARS BEANS ARE
- OPĆINA PODGORAČ TRG P PEJAČEVIĆA 2 31 433 PODGORAČ
- ASIGNACIÓN PLANIFICAR UNA ASIGNATURA Y DESARROLLAR UNA UNIDAD DIDÁCTICA
- PRZEDMIOTOWE ZASADY OCENIANIA OBOWIĄZUJĄCE NA LEKCJACH CHEMII I PRACE
- EΛΛΗΝΙΚΗ ΔΗΜΟΚΡΑΤΙΑ ΥΠΟΥΡΓΕΙΟ ΠΑΙΔΕΙΑΣ ΠΆΤΡΑ 02122011 ΔΙΑ ΒΙΟΥ ΜΑΘΗΣΗΣ
- D SAUPOO STAKEHOLDERS PARA PROVEER PRODUCTOS YO SERVICIOS DE
- FS RASKOG OKRUGA SS ORGANIZACIJA SUDIJA TEHNICKA SUDIJSKA KOMISIJA
- ACP WGN04 NEWORLEANS LIST OF PARTICIPANTS WORKING
- 2011 DIRECTORY OF STATE SUPERINTENDENT ASSOCIATIONS COMPILED FROM PUBLICLYAVAILABLE
- PLAN DE TRABAJO DE LA BECA ESTIMULO 1
- FIVB VOLLEYBALL AT SCHOOL SYMPOSIUM PAGE 4 OF 4
- Lectura 5 la Universidad y la Muerte de Franco
- A REVIEW ON BIG DATA ANALYTICS VVADIVU ASSISTANT PROFESSOR
- ANEXO N°3 CONSENTIMIENTO INFORMADO DEL ESTUDIO CONSENTIMIENTO INFORMADO PARA
- THE MEADOWS SCHOOL – TERM DATES 202122 AUTUMN TERM
- FIRECUPA 360 WEST SECOND ST OXNARD CA
- ZP23801312910772014 KATOWICE 11042014 R EGZ POJEDYNCZY KOMENDA WOJEWÓDZKA POLICJI
- PROJECT AND PROCEDURAL RISK ASSESSMENT LONDON CENTRE FOR NANOTECHNOLOGY
- LIMPOPO RURAL TRANSPORT STRATEGY VHEMBE DISTRICT – BENDE MUTALE
- 11 PATVIRTINTA LIETUVOS GEOLOGIJOS TARNYBOS PRIE APLINKOS MINISTERIJOS DIREKTORIAUS
- FORMULARIO DE POSTULACIÓN – LEY N° 20996 CARTA DE
- 66 HVIEZDOSLAVOV KUBÍN – HARMONOGRAM POSTUPOVÝCH SÚŤAŽÍ I
- 114 LIETUVOS RESPUBLIKOS VYRIAUSYBĖ NUTARIMAS DĖL LIETUVOS RESPUBLIKOS VALSTYBĖS
- CONVENIO DE COLABORACIÓN ENTRE LA UNIVERSIDAD DE CÁDIZ LA
- HOLA MARTA POTS DONAR CÒPIA DE LES LLETRES
- DIRECCION GENERAL DEL ORGANISMO DE CUENCA FRONTERA SUR DIRECCION
- G O S P O D A R K
CONSUMER DIRECTED SERVICES SERVICE REPORT INSTRUCTIONS SECTION I
DEPARTMENT OF HOMELAND SECURITY US COAST GUARD AUXILIARY 11SR
 S EMESTER 2 CHAPTER 9 EIGRP V 40 901
S EMESTER 2 CHAPTER 9 EIGRP V 40 901“LAS PALABRAS COMO CONSTRUCCIONES ALGUNAS REFLEXIONES A LA LUZ
 HEALTHY CAMPUS 2020 – STUDENT OBJECTIVES | 0 H
HEALTHY CAMPUS 2020 – STUDENT OBJECTIVES | 0 HPRZEDMIAR ROBÓT REMONT DROGI GMINNEJ NR I92 W MSC
 WAT053372 REV 7 PERFORMANCE MAINTENANCE PROTOCOL 99629962998 PDA
WAT053372 REV 7 PERFORMANCE MAINTENANCE PROTOCOL 99629962998 PDASPECJALNOŚĆ ZARZĄDZANIE ŚRODOWISKIEM SERWEROWYM PRZEDSIĘBIORSTW BEZPIECZEŃSTWO INFORMACJI W SYSTEMACH
 DUE DILIGENCE CERTIFICATE THIS IS TO CERTIFY THAT THE
DUE DILIGENCE CERTIFICATE THIS IS TO CERTIFY THAT THEREFERENCE DOCUMENT A GREATER MANCHESTER CHURCHES TOGETHER REGISTERED CHARITY
 POLIZIA PROVINCIALE PIANO DI CONTROLLO FAUNISTICO CON FINALITA’
POLIZIA PROVINCIALE PIANO DI CONTROLLO FAUNISTICO CON FINALITA’ZÁKLADNÍ ŠKOLA A MATEŘSKÁ ŠKOLA HORNÍ BRADLO OKRES CHRUDIM
NORMAS PARA LA PRESENTACIÓN DE COMUNICACIONES A CONDICIONES PARA
RECIBO DE ENTREGA DE CESTA BÁSICA RAZÃO SOCIAL
 SLOŽENÍ POROT CENY MINISTERSTVA KULTURY ZA PŘÍNOS V OBLASTI
SLOŽENÍ POROT CENY MINISTERSTVA KULTURY ZA PŘÍNOS V OBLASTI 5 PROVISIONS LIST CLIENT’S NAME CHARTER CODE CHARTER PERIOD
5 PROVISIONS LIST CLIENT’S NAME CHARTER CODE CHARTER PERIOD C LINICA UNIVERSITARIA CENTRO DE INFORMACIÓN DE MEDICAMENTOS SERVICIO
C LINICA UNIVERSITARIA CENTRO DE INFORMACIÓN DE MEDICAMENTOS SERVICIODC45A (919) PAGE 2 OF 2 PROFESSIONAL SERVICES NEW
 6 HRVATSKA PSIHOLOŠKA KOMORA NA TEMELJU ČLANKA 25 ZAKONA
6 HRVATSKA PSIHOLOŠKA KOMORA NA TEMELJU ČLANKA 25 ZAKONAPROPOZICIJE 2018 PRVENSTVA PLIVAČKOG SAVEZA VOJVODINE ZAJEDNIČKE ODREDBE ČLAN