PEXSERIES OF PRECISIONSHUTTLE DESTINATION VECTORS FOR PROTEIN EXPRESSION IN
PEXSERIES OF PRECISIONSHUTTLE DESTINATION VECTORS FOR PROTEIN EXPRESSION IN
OTI Letterhead
pEX-series of PrecisionShuttle Destination Vectors
For Protein Expression in E.coli
pEX-N-His-GST
pEX-N-GST
pEX-N-His
pEX-C-His
pEX-1
Introduction
The pEX-series of vectors are destination vectors for protein expression in E. Coli. The T7 promoter in the vector allows the encoded protein to be expressed in BL21(DE3) cells upon IPTG induction. The pEX-series of vectors also provide a TEV (the tobacco etch virus) protease cleavage site to remove any of the tags after protein purification. The pEX N-terminal tagged vectors share the same multiple cloning sites as other destination vectors to facilitate easy transfer of the ORF region from any of OriGene’s standard TrueORF clones in the pCMV6-Entry vector. pEX-1 and pEX-C-His vectors have a BseR I site instead of Sgf I due to the restriction of E. Coli RBS (ribosome-binding site) for higher protein expression. BseR I and Sgf I produce compatible sticky ends, so pEX-1 and pEX-C-His vectors are still compatible with transferring the ORF region from any of OriGene’s standard TrueORF clones in the pCMV6-Entry vector or other destination vectors. The PrecisionShuttle, a one-step cut/paste procedure, is fast, reliable, and cost effective. The different antibiotic resistance marker in the destination vectors (Ampicillin instead of Kanamycin in the Entry Vector) is designed for easy screening of the successfully shuttled plasmids. The diagram below shows how to do subcloning from TrueORF Entry into other destination vectors.
Note: to clone an ORF into pEX-1 and pEX-C-His vectors, BseR I/Mlu I will be used instead of the typical Sgf I /Mlu I.
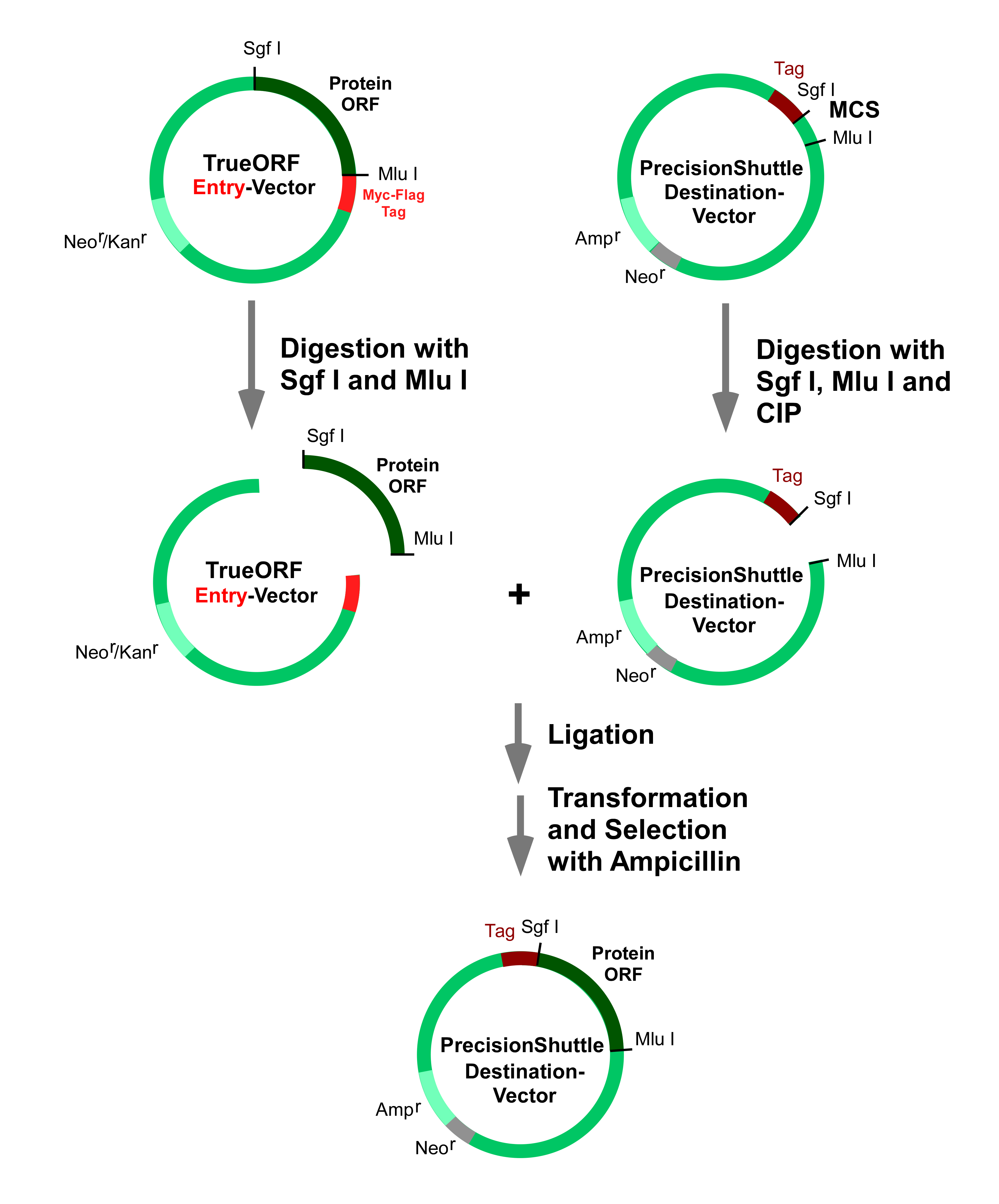
The pEX vector series have several tagging options, tagged and non-tagged or on either the N- or C-terminus. Depending on the desired purification strategy or desired fusion partner, one can choose the N-His-GST-, N-GST, N-His-, C-His-tagged or non-tagged bacterial expression vector. For details, please see http://www.origene.com/cdna/TrueORF/DestinationVector.aspx , then click on the “Bacterial expression” tag. Inducible expression of the gene of interest is directed by the hybrid T7/lacO promoter in the presence of isopropyl-β-D-thiogalactopyranoside (IPTG) in the cultured media, whereas in the absence of IPTG, a copy of the Lac repressor gene (lacI) carried in this vector mediates a tight repression of the protein expression. The RBS is located prior to the ORF insert to promote efficient and accurate translation of mRNA in bacteria. The affinity tag linker contains a TEV protease cleavage site that provides a convenient option for the removal of the fusion tag(s) from the protein of interest. Although in many cases, the presence of a GST or His tag does not interfere with the normal function of the target protein. However, if tag removal is needed, the TEV protease offers a highly specific cleavage option that is effective in a broad range of temperatures and salt concentrations. All pEX- expression vectors carry the ampicillin resistance selection marker.
5’ sequence primer
VP1.5 GGACTTTCCAAAATGTCG for all pEX-non-GST vectors
GST_F AACGTATTGAAGCTATCCCAC for pEX-N-His-GST and pEX-N-GST
3’ sequence primer
XL39 ATTAGGACAAGGCTGGTGGG
Table 1: Feature locations in the pEX vector series
|
Feature |
pEX-N-His-GST |
pEX-N-GST |
pEX-N-His |
pEX-C-His |
pEX-1 |
|
|
|
|
|
|
|
|
T7 promoter |
217-236 |
217-236 |
217-236 |
217-236 |
217-236 |
|
Lac Operator |
231-263 |
231-263 |
231-263 |
231-263 |
231-263 |
|
Ribosome binding site |
279-288 |
279-288 |
279-288 |
279-288 |
279-288 |
|
6x-His tag |
295-315 |
- |
288-308 |
359-376; 416-433 |
- |
|
GST tag |
322-981 |
295-954 |
- |
- |
- |
|
TEV site |
997-1017 |
970-990 |
309-329 |
338-358; 395-415 |
- |
|
MCS |
1018-1104 |
991-1077 |
330-416 |
288-449 |
278-364 |
|
T7 Terminator |
1129-1189 |
1102-1162 |
441-501 |
474-534 |
389-449 |
|
LacI repressor coding seq. |
1513-2595 |
1486-2568 |
825-1907 |
858-1940 |
773-1855 |
|
ColE1 ori. of replication |
2988-3661 |
2961-3634 |
2311-2973 |
2333-3006 |
2248-2921 |
|
Amp. resist. coding seq. |
3805-4665 |
3778-4638 |
3128-3977 |
3150-4010 |
3065-3925 |
Protocols
Transfer an ORF from a TrueORF Entry vector into a pEX destination vector
To transfer the protein-coding region from a TrueORF Entry Vector (donor) to a
pEX-vector (recipient), please follow the transfer protocol below *.
1. Digest the TrueORF entry clone:
Component Volume
10X restriction buffer** 2 μl
Sgf I (10 U/μl) 0.6 μl
Mlu I (10 U/μl) 0.6 μl
nuclease-free water 13.8 μl
TrueORF in pCMV6-Entry (100-200ng) 3 μl
--------------------------------------------------------------------------
Total volume 20 μl
Incubate at 370C for 2 hours.
2. Digest the TrueORF destination vector:
Component Volume
10X restriction buffer** 2 μl
Sgf I (10 U/μl) 0.6 μl
Mlu I (10 U/μl) 0.6 μl
nuclease-free water 14.8 μl
Selected Destination vector (200ng) 2 μl
---------------------------------------------------------------------------
Total volume 20 μl
Incubate at 37C for 2 hours. Add calf intestine phosphatase to the digestion, and continue to incubate at 37C for an additional 30 minutes.
* For the 4% of the transcripts that have internal Sgf I or Mlu I sites, please use the appropriate combination of restriction sites as recommended by OriGene.
Special instruction for transferring ORF into the two E. Coli expression vectors, pEX-1 (cat# PS100075) and pEX-C-His (cat# PS100076). Sgf I site is not available in the MCS of these two vectors. These two vectors need to be cut with BseR I and Mlu I instead of Sgf I and Mlu I for ORF subcloning. However, the ORF insert from the TrueORF entry vector can still be cut with Sgf I and Mlu I. PS100075 and PS100076 cut with BseR I have compatible sticky ends with ORF insert cut with Sgf I. PS100075 and PS100076 vector backbones digested with BseR I and Mlu I can be ligated with ORF inserts digested with Sgf I and Mlu I.
** Sgf I/ASIS I from Fermentas works better.
3. Purify the digestion using a commercial PCR purification column and elute in 20 ul 10mM Tris.
4. Set up a ligation reaction:
Component Volume
10 x T4 DNA ligation buffer 1 μl
T4 DNA Ligase (4U/μl) 0.75 μl
nuclease-free water 3.25 μl
digested DNA from Step 1 2 μl
digested DNA from Step 2 3 μl
---------------------------------------------------------------------------
Total volume 10 μl
Incubate the ligation reaction at room temperature for 1 hour. (Optimal temperature for some larger inserts may be around 15oC)
5. Transform the ligation reaction into the high-efficiency, competent E. coli cells (≥ 1×108 CFU/μg DNA) following the appropriate transformation protocol. Plate the transformants on LB-agar plates supplemented with 100 μg/ml ampicillin.
6. Pick at least four colonies for subsequent DNA purification and screening. Amplify and purify the selected clones by growing overnight in liquid LB-amp media, then isolating the DNA using standard plasmid purification procedures, http://www.origene.com/Other/Plasmid_Purification/ .
7. Confirm the insert by restriction digestion and/or vector primer sequencing using the provided VP1.5 for 5’ end sequencing and XL39 for 3’ end sequencing.
Subcloning an ORF (other than OriGene TrueORF) into a pEX- expression vector
If you have a gene of interest in a non-OriGene vector and wish to clone the ORF into any of OriGene’s destination vectors, please follow the protocol listed in OriGene’s TrueORF clone application manual http://origene.medigent.com/assets/Documents/TrueORF/TrueORFApplicationGuide.pdf in the sections, “The Primer Design and PCR Amplification of ORF” and “Cloning of ORF into the Entry Vector”. Briefly, the rare cutters, SgfI and MluI sites, are appended to PCR primers in the correct orientation for insertion into pEX vectors digested with the same or BseR I/Mlu I.
Transformation into bacterial (expression host) competent cells
The pEX-ORF plasmids should be transformed into an appropriate expression host cell, such as BL21 (DE3), using standard transformation protocols from the competent cell manufacturer. Plate 5% of the transformants on LB-ampicillin agar plates.
Protein Expression and Solubility (induction studies)
To test the fusion protein’s expression yield and solubility, select a colony from the ampicillin plate and grow in ampicillin LB media overnight. Dilute 0.2 ml of the overnight culture into 2.0 ml of the fresh media and incubate for 1 hr at 37C. Protein expression is induced with Isopropyl-1-thio-β-D-galactopyranoside (IPTG) (50 μM–1000 μM) for a variable period (2-24 hrs) at 37C or lower temperature. Simultaneously, incubate a control sample without IPTG. At time points between 2 and 24 hours, remove 0.1 ml culture, spin down and collect pellet. Add 0.1 ml of the lysis buffer (recipe below) and rock at room temperature for 20 min. Create a soluble fraction by centrifuging at 14,000 x g at 4C for 20 minutes. Add an appropriate amount of SDS loading buffer to both soluble and insoluble fractions and save at –20C. After all of the time points are collected, load 25-50 ul of both fractions from all time points on an SDS-PAGE gel for Coomassie Blue staining analysis and/or western blot analysis.
Lysis Buffer:
20 mM Tris, 150 mM NaCl, 1% NP40, 1 mM NaF, 1 mM PMSF, pH 7.4
TEV Proteolytic Cleavage to remove the fusion tag
For optimal cleavage and removal of cleavage products, please follow the manufacturer’s protocol. OriGene has tested and recommends using the TEV protease from Promega (Promega, V6051. http://www.promega.com/catalog/catalogproducts.aspx?categoryname=productleaf_1712 ).
Tags: destination vectors, origene’s destination, expression, vectors, protein, precisionshuttle, pexseries, destination
- FILED 4521 CERTIFIED FOR PUBLICATION IN THE COURT OF
- MẪU SỐ 13ĐK CỘNG HÒA XÃ HỘI CHỦ NGHĨA
- SYSTEMS CHANGE APPENDIX A WISE & HEALTHY AGING LONGTERM
- POWERPLUSWATERMARKOBJECT24491627 ANNEXE L8 FICHE D’IDENTIFICATION DU CONCURRENT «
- ACTIVIDADES PROPUESTAS PARA LA SEMANA DEL 11 AL 15
- EKSEMPEL PÅ PROSJEKTPLAN DET PRESISERES AT DETTE DOKUMENTET ER
- KRITERIJI ZA KVALITATIVNI ODABIR PONUDITELJA 1 NARUČITELJ JE
- PREMIOS A LA EXCELENCIA EN RECURSOS HUMANOS DE
- STRATEGIC PLAN LEVEL RIVERSIDE PRIMARY SCHOOL 201516 COBB
- APPENDIX 6 HOUSING SUPPLY TARGET METHODOLOGY 10 INTRODUCTION 11
- E NNA ……………………………………………………… VEÍ DE AMB DOMICILI A
- META PUGLIA PERIODO DAL 27 DICEMBRE’03 AL 02
- REPUBLIKA SRBIJA PREKRŠAJNI SUD U MLADENOVCU UPBRSU M L
- PREFERRED WORKER JOB OFFER LETTER SEE OAR 4361100290(2) FOR
- KAJIAN PSIKOLOGI LINTAS BUDAYA DALAM LINGKUP SELFKEPRIBADIAN A PENDAHULUAN
- TERCERA FILA DE ASIENTOS SU VEHÍCULO AUDI Q7 CAMPAÑA
- SOL·LICITUD DE FACTURA PER A TÍTOLS PROPIS SOLICITUD DE
- CAMINO REAL DE TENNESSEE WILLIAMS ADAPTACIÓN DE NURIA ALKORTA
- MARIE CURIE JOB DESCRIPTION JOB TITLE REGULAR GIVING MANAGER
- FONÉTICA (RESUMEN)CURS00910 DEL LATÍN A LAS LENGUAS ROMANCES LAS
- CN17729 EFFECTIVENESS AND EFFICIENCY OF REGULATORY BODIES A MEXICAN
- MEDICAL RECORDS LABEL ALERT RESEARCH PROJECT ALERT RESEARCH PROJECT
- G OBIERNO DE NAVARRA CIRCUITO DE TRAMITACIÓN DATOS DEL
- 1)EKMEK LIMON GIBI BESINLERIN ÜZERINDE KÜF OLUŞTURAN CANLILAR AŞAĞIDAKILERDEN
- EJERCICIOS TEMA 5 EL CIERRE DE LA CONTABILIDAD Y
- UNIVERSIDADE FEDERAL DE PELOTAS FACULDADE DE ODONTOLOGIA PROGRAMA DE
- ProfessorWillyToreMrch
- CONTENIDOS 1º PRIMARIA MATEMÁTICAS BIENVENIDOS A PRIMERO WEB INTERACTIVAS
- CRISIS SUPPORT SCALE (JOSEPH ET AL 1992) WE ARE
- MEMÒRIA PER A LA PARTICIPACIÓ EN LA CARTERA DE
 DEN NORSKE KYRKJA ETNEDAL KYRKJELEGE FELLESRÅD MØTEPROTOKOLL NR 22016
DEN NORSKE KYRKJA ETNEDAL KYRKJELEGE FELLESRÅD MØTEPROTOKOLL NR 22016LISTA NAGRODZONYCH W KONKURSIE LITERACKIM URZEKŁA MNIE OSTROWSKA ZIEMIA
FICHA TÉCNICA 1 NOMBRE DEL MEDICAMENTO GLIZOLAN 50 MG
MINISTERIO DE JUSTICIA COMISIÓN GENERAL DE CODIFICACIÓN SECCIÓN ESPECIAL
 PROGRAMA PRELIMINAR AULA MAGNA DEL HOSPITAL JUAN A FERNÁNDEZ
PROGRAMA PRELIMINAR AULA MAGNA DEL HOSPITAL JUAN A FERNÁNDEZOFSAA METRO REGION MEET HYTEKS MEET MANAGER PAGE
WWWPUNTAJECOMAR – SITIO WEB DOCENTE ART 67 RESOLUCIÓN Nº
 WOMEN’S POLICY PLATFORM WOMEN COMPRISE OVER 50 PERCENT OF
WOMEN’S POLICY PLATFORM WOMEN COMPRISE OVER 50 PERCENT OF CONSEJOS PARA EL DESARROLLO DE SU CARRERA 1 ARMONICE
CONSEJOS PARA EL DESARROLLO DE SU CARRERA 1 ARMONICE 2400 VIA CANELA OROVILLE CA 95966 (530) 5326000 DIRECTORA
2400 VIA CANELA OROVILLE CA 95966 (530) 5326000 DIRECTORA5 THE ENVIRONMENTAL STUDIES INTERNSHIP NOVEMBER 2020 A GOOD
VAI TRÒ CỦA VẠN CHÀI QUẢN LÝ NGHỀ CÁ
 POWERPLUSWATERMARKOBJECT3 A FILM REVIEW TASK PICK A FILM THAT
POWERPLUSWATERMARKOBJECT3 A FILM REVIEW TASK PICK A FILM THATEVALUATION A L’ENTREE EN SECONDE SEMUR CHATILLON –
CASO PRÁCTICO LA DEMANDA DERECHO PROCESAL CIVIL SUPUESTO DE
 CONTRATO PARA LA REALIZACIÓN DEL ESTUDIO POSTAUTORIZACIÓN CON PRODUCTO
CONTRATO PARA LA REALIZACIÓN DEL ESTUDIO POSTAUTORIZACIÓN CON PRODUCTOSPECYFIKACJA ISTOTNYCH WARUNKÓW ZAMÓWIENIA POSTĘPOWANIE PROWADZONE W TRYBIE
3 EQUITY STRATEGIES 20032006 AS APPROVED BY SENATE ON
DIALOGUE WITH NATIONAL COMMITTEE ON USCHINA RELATIONS BY YANG
 CURSO CURSO DE AUTODEFENSA PSÍQUICA PARA TENER CUIDADO DE
CURSO CURSO DE AUTODEFENSA PSÍQUICA PARA TENER CUIDADO DE